|
|
 |
Calcium signalling
Contact: Geneviève Dupont
Collaboration: Laurent Combettes (Paris-Sud)
Intracellular calcium dynamics is highly organized in time and space. We focus on the molecular mechanisms responsible for this organization, which allows calcium to serve as a universal second messenger. Particular emphasis is put on the role of cellular calcium signalling in health and disease.
|
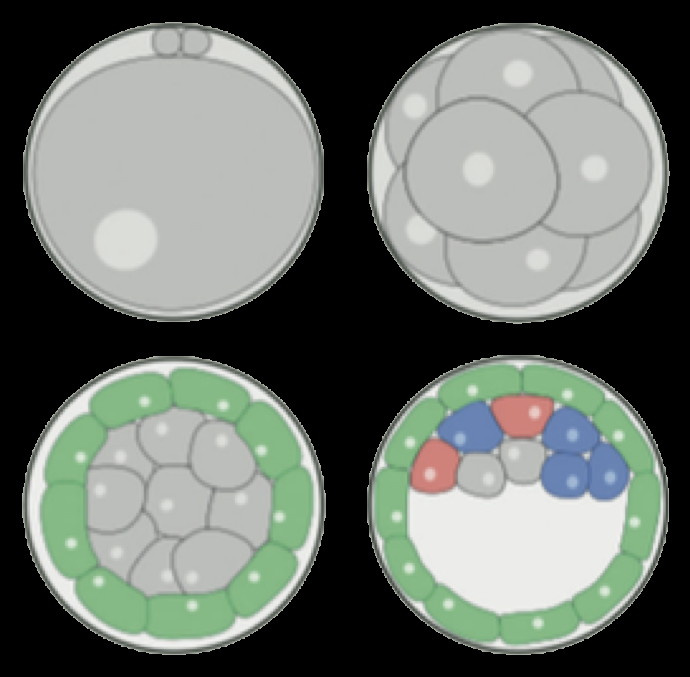 |
Modeling embryonic cell fate determination
Contact: Geneviève Dupont
Collaboration: Claire Chazaud (Clermont-Ferrand)
During early murine embryogenesis, cells from the inner cell mass (ICM) can be specified in epiblast (Epi, future embryonic tissues) or primitive endoderm (PrE, future placenta and other extra-embryonic tissues). The specification is controlled by a gene regulatory network, involving Erk signaling through extracellular Fgf4. We have developed a model describing the specification process in terms of tristability, bifurcation diagrams, and inter-cellular coupling through the secretion and diffusion of Fgf4.
|
 |
Modeling the plant circadian clock
Contact: Jean-Christophe Leloup and Didier Gonze
Collaboration: Christian Hermans and Nathalie Verbruggen (ULB)
The plant circadian clock is particularly complex. It involves several clock genes, expressed at specific times of the day, multiple regulatory feedback loops, and multiple light entries. We are developing a compact model that recapitulates these regulations and use it to understand the entrainment by light-dark cycles as well as the dynamics of clock-regulated processes.
|
 |
Computational analysis of biological clocks design
Contact: Didier Gonze
Collaboration: Thomas Erneux (ULB)
We analyze the dynamics of biomolecular oscillatory systems by means of mathematical and numerical analyses. We also study synchronization and entrainment properties of oscillations and relate their feature (e.g. damped vs self-sustained, delay, non-linearity) with entrainment performance and robustness to parameter changes.
|
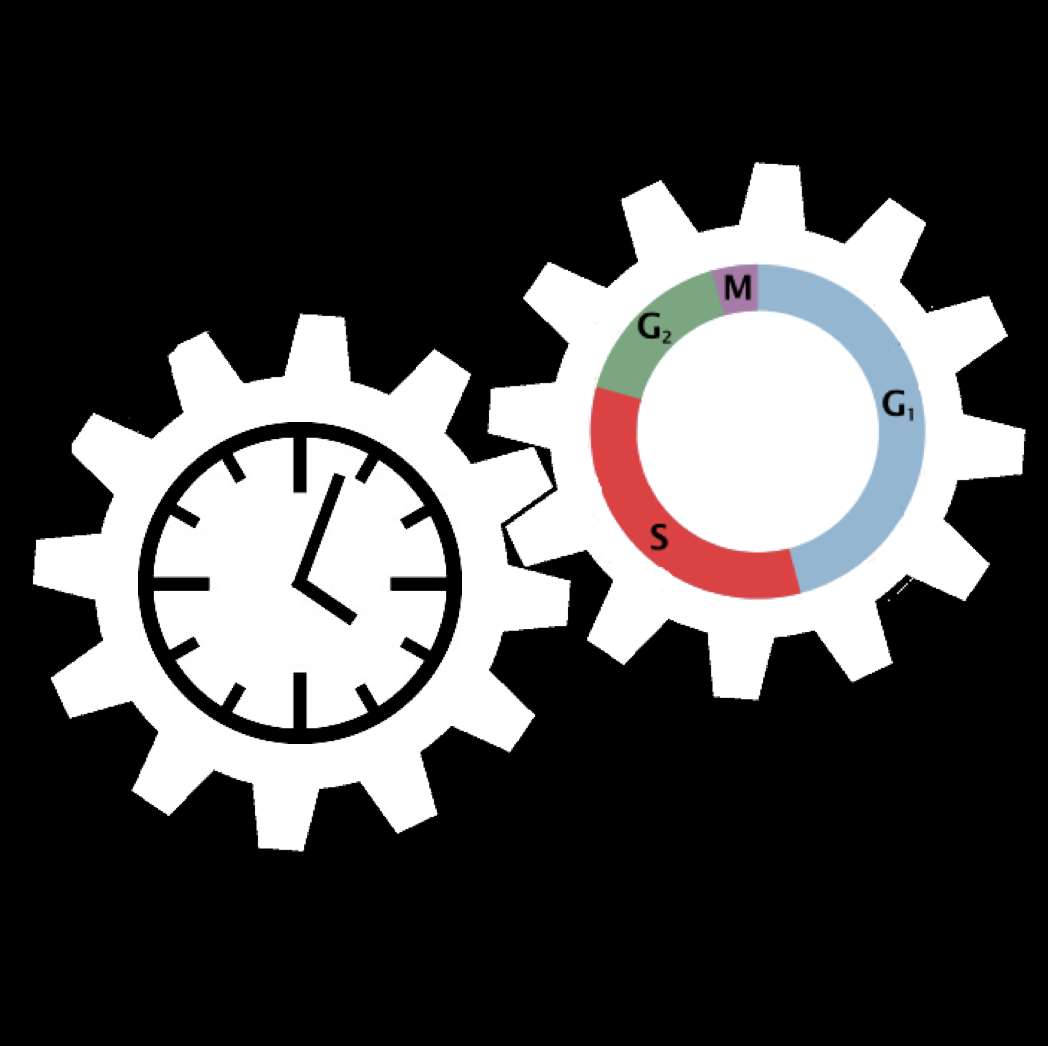 |
The mammalian cell cycle and the circadian clock : Modeling the dynamics of two coupled cellular rhythms
Contact: Albert Goldbeter
Collaborations: Jie Yan (Soochow University, Suzhou, China), Claude Gérard (De Duve Institute, UCL, Brussels), Bert Van der Horst (Erasmus University, Rotterdam)
The cell cycle and the circadian clock represent two major cellular rhythms. These two oscillatory networks are coupled through multiple regulatory interactions. Our goal is to determine by means of detailed mathematical models based on experimental observations how the coupling of the two networks affects their oscillatory dynamics.
|
 |
Modeling the dynamics of microbial communities
Contact: Didier Gonze
Collaborations: Karoline Faust (KU Leuven), Jeroen Raes (KU Leuven), Leo Lahti (KU Leuven / Turku, Finland), Isabelle George (ULB)
The composition of microbial communities is dynamic: The relative abundances of species within a community vary with time and change with environmental conditions. We are developing models to study the response of such microbial communities to environmental perturbations or to explain the emergence of various community types.
|
|

